

- Clc bio genomics workbench manual mac os x#
- Clc bio genomics workbench manual driver#
- Clc bio genomics workbench manual windows 10#
- Clc bio genomics workbench manual download#
- Clc bio genomics workbench manual torrent#
Thus a large deviation from the ideal value and a small tolerance will give a large negative contribution in the final score and a small deviation from the ideal and a high tolerance will give a small negative contribution in the final score. The negative contribution to the final score is determined by how much the parameter deviates from the ideal value and is scaled by the specified tolerance:Ĭontribution = (ideal - actual value)/tolerance
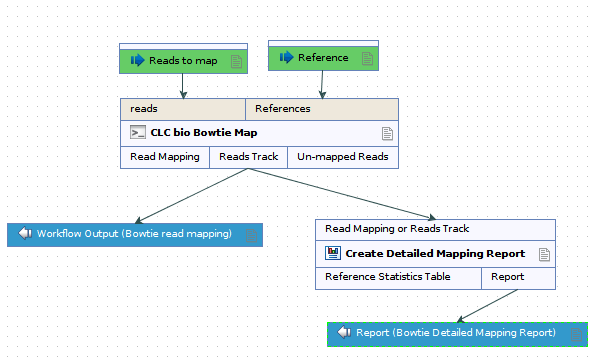
Self-annealing score: the ideal value is 0 and the tolerance corresponds to the maximum value specified in Each parameter is assigned an ideal value and a tolerance. The ideal score for a pair is 100 and pairs are ranked inĭescending order. Primer pairs such as the pair-annealing score. Parameters related to individual primers, such as the secondary structure score, and parameters related to By CLC bio CLC Genomics Workbench (64-bit) for analyzing and visualizing Next Generation Sequencing data, incorporates cutting-edge technology and algorithms, while also supporting and integrating. WholeGenomeAlignment : WholeGenomeAlignment.cpaīGA 20.0.2 : BiomedicalGenomicsAnalysis.CLC Main Workbench employs a proprietary algorithm to rank primer pairs. Genome Finishing Module 21 : CLCGenomeFinishingModule_21.cpaĪdditionalAlignments : AdditionalAlignments.cpa Long Read Support 21 : LongReadSupport_21.cpaĪdditional Alignments 21 : AdditionalAlignments_21.cpaĪnnotate with GFF file 21 : AnnotatewithGFFfile_21.cpa Whole Genome Alignment 21 : WholeGenomeAlignment_21.cpa Microbial Genomics Module 21 : CLCMicrobialGenomicsModule_21.cpaīiomedical Genomics Analysis 21 : BiomedicalGenomicsAnalysis_21.cpa

Local BLAST CLC_Genomics_Workbench_Local BLAST.pdf
Clc bio genomics workbench manual mac os x#
A discrete graphics card from either Nvidia or AMD/ATI.
Clc bio genomics workbench manual driver#
Please make sure the latest driver for the graphics card is installed
Clc bio genomics workbench manual download#
Minimum linux versions needed for the SRA download functionality: Red Hat 6, Fedora 12, and SUSE 11.
Clc bio genomics workbench manual windows 10#
Clc bio genomics workbench manual torrent#
Sanger, Roche, Applied Biosystems, Illumina, Helicos BioSciences, Ion Torrent 등에서 생성된 멀티 포맷 데이터의 분석을 할 수 있습니다. * Chromatin immunoprecipitation sequencing (ChIP-seq) 분석 * MA-와 boxplots으로 구성된 분석 Quality control 툴 * Heat map, table, scatter plot view를 통한 시각화 * 일반적인 RNA-seq 분석 (Gene expression profile) * mRNA sequencing 데이터의 유전자 발현 calculation * Large scale의 mutation과 rearrangement 그래픽 툴 * Standard read, paired-end read, mate pair read 지원


 0 kommentar(er)
0 kommentar(er)
